Transfer of Escherichia Coli
Transfer of Escherichia Coli
By: Amudha Venugopalan, Sarah Councilor, and David Roehrich
Introduction:
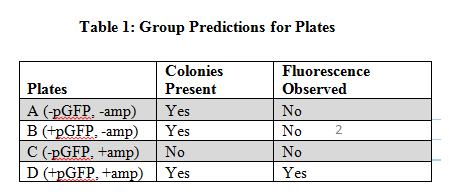
Escherichia coli are prokaryotic bacteria that can be transformed by lateral gene transfer. E. coli are transformed by direct transfer which means the bacterial cell directly uptakes the DNA of the plasmid introduced to it (Wimpee et al. 2013). A plasmid is a circular small piece of self-replicating DNA, plasmids carry fewer genes therefore DNA can be passed between species of bacteria (Wimpee et al. 2013). The plasmid we introduced to the E. coli is the plasmid pGFP, this plasmid codes for Green Fluorescent Protein (GFP) and Ampicillin Resistance (AMPr). Plasmid transfer of DNA into bacteria like E. coli can create antibiotic resistance in bacteria. This is important for understanding how bacteria evolve and how humans can treat bacteria that causes disease (Miroux et al. 1966). We use pGFP so that we can visually see if the plasmid DNA transfer into E. coli occurred. The Green Fluorescent Protein is visible in UV light so if the bacteria do transform then we know they will be antibiotic or ampicillin resistant as well. For each plate we had different independent and dependent variables. For plates A and C the independent was the presence of ampicillin which plate C had and plate A did not, but both plates did not contain the GFP plasmid which was the dependent variable. For plates B and D the independent variable was the presence of ampicillin as well which plate D did have and B did not, but both plates contained the GFP plasmid. We predicted that plates not containing pGFP will have colonies on plates without ampicillin, while plates with pGFP will have colonies present on plates with ampicillin (Table 1). We hypothesized that transformed bacteria colonies will be present on agar plates with ampicillin and without ampicillin.
Methods
We used four different agar plates in this experiment: plate A (no plasmid DNA or ampicillin), plate B (plasmid DNA, no ampicillin), plate C (no plasmid DNA, ampicillin present), and plate D (plasmid DNA and ampicillin present) (Wimpee et al. 2013). To start the experiment we treated the E. coli cells with Calcium Chloride (CaCl2), which helps the thick cell wall of E. coli break down and become more permeable to the pGFP DNA (Wimpee et al. 2013). We then mixed the DNA with the E. coli Calcium Chloride treated cells, incubate and heat-shock them before adding them to plates.
Procedure:
- Use two separate tubes containing 50 microliters of transformable E. coli cells. One tube will have no plasmid DNA (this is the control) and number it 1. The second tube we will use as the experimental E. coli and number 2.
- Add 5 microliters of pGFP DNA into the tube labeled 2. Then put both of the tubes on an ice bath for 20 minutes.
- While the cells are incubating, label four LB agar plates A,B,C,D.
- A (-pGFP,-amp)
- B (+pGFP,-amp)
- C (-pGFP,+amp)
- D (+pGFP,+amp)
- When the test tubes are done on the ice bath move them to a water bath at 42 degrees Celsius for only one minute.
- When finished move the tubes to the ice bath again for only two minutes.
- Pipet 500 microliters of NZY recovery medium into both test tube 1 and 2.
- Then place the test tubes into a water bath of 37 degrees Celsius for 60 minutes.
- After the 60 minute wait, pipet 100 microliters of test tube 1 onto agar plate A and C. Then pipet 100 microliters of test tube 2 onto plates B and D.
- Place 5 sterile glass beads onto the plate after placing the bacterial cells on and swirl around slowly on the plate to spread the bacterial cells.
- Place the plates into an incubator at 37 degrees Celsius for 12-16 hours.
- Then place the plates into a refrigerator until ready to view results.
- When plates are ready to be view place plates under a UV light to see if the growing colonies have transformed.
(Wimpee et al. 2013)
Results
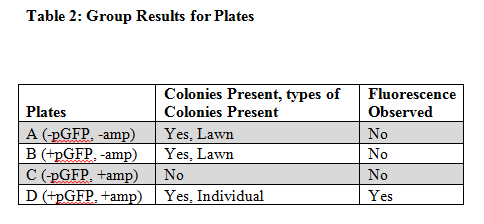
Plate A which contained no plasmid DNA and no ampicillin had colonies of bacteria grown on it and had no observable fluorescence. Plate B which contained plasmid DNA and had no ampicillin had colonies of bacteria grown on it and had no observable fluorescence. Plate C which had no plasmid DNA and had ampicillin present had no colonies grown on it and no observable fluorescence. The Final plate D which had plasmid DNA and had ampicillin present had colonies of bacteria present and it had observable fluorescence. Plate A, B, and C had colonies present and plate A and B shared colony growth in a similar lawn pattern, meaning it spread abundantly all over the plate. Whereas plate C had individual colony growth and was less abundant in its expansive growth.
Discussion
The results of this experiment supported our hypothesis and predictions (Table 2). The class results were also the same as ours. The plates that had no ampicillin present contained the transformed E. coli because the plasmid pGFP contained ampicillin resistance (AMPr) which codes for the ability to be resistant to plates containing ampicillin. Since plates A and B both did not have ampicillin this wouldn’t matter. The bacteria would’ve grown if it were transformed or not since it didn’t have antibiotics in it, but plate D had ampicillin on the plate and we saw transformed bacterial growth because the plasmid was present and created resistance to the ampicillin. Plate C did not show growth because it contained ampicillin and we put test tube one’s sample of E. coli on it which did not contain the plasmid pGFP, meaning it would not be transformed and then could not grow on ampicillin. In addition we did not see any fluorescence on plate A because there was no plasmid DNA which made the Green Fluorescent Protein not transformed into the bacteria, this same principle applies to plate C which had no plasmid DNA. Plate B contained plasmid DNA and could have possibly had fluorescence visible but we didn’t see much because the colonies are not selected for. The plasmid contained E. coli require more energy to show fluorescence. Therefore growth of the transformed E. coli was very low. As for plate D, Table 2 shows that it showed both colonies and fluorescence. The fluorescence were observable because plate D contained the transformed E. coli bacteria. The evidence of these bacteria’s transformation are important for the research of bacteria transformations so we can better learn bacteria’s ability to adapt to different environments (Roberts et al. 1955). When we put this plasmid in E. coli we can learn how bacteria similarly can be transformed to prevent diseases and illness in humans and also help create new forms of organic resources and ecosystems for much of the planet (Anderson 1966).
Literature Cited
Wimpee, C.F., O. Alminas, E. Muslin and S. Hoot. 2013. BIO SCI 152 Laboratory 2: Gene Transfer in Escherichia coli. Available from D2L.
Anderson, E. S. 1966. Possible Importance of Transfer Factors in Bacterial Evolution. Pages 637-638 in Nature. Available from web.
Roberts, Richard B., D. B. Cowie, P. H. Abelson, E. T. Bolton, and R. J. Britten. 1955 Studies of Biosynthesis in Escherichia Coli Vol X. In Washington: Carnegie Institution of Washington. Available from web.
Miroux, Bruno, and John E. Walker. 1966 Over-production of Proteins in Escherichia Coli: Mutant Hosts That Allow Synthesis of Some Membrane Proteins and Globular Proteins at High Levels. Pages 289-298 in Journal of Molecular Biology 260.3. Available from web.
