High-Efficiency Transformation of Yeast
Written by Isreal
Introduction:
The CDC42 gene or Cell Division Control 42 gene has homologies in many species ranging from yeast to humans. This gene controls and influences many pathways that are important in the formation of many proteins. One of the great things about CDC 42 is that we can use a mutated strain and, through transformation, actually change the genetic material within the cell (Chapoel, et al 2010). This would cause complementation within the cell. Transformation is the exchange or uptake of genetic material, which ultimately changes the genetics of any given cell.
Transformation is being wildly studied because it could result in many cures and breakthroughs in the medical field. An example of this transformation would be injecting fish DNA into plants to allow them to function at cooler temperatures. This is helpful because useful traits from a given organism can now be expressed in other cells and organisms that lack the trait (Ivics, et al 1997). In this lab in particular, we are looking at the transformation of a cdc42ts strain of yeast. This strain has a mutation in the CDC42 gene that causes the cells to be unable to grow at higher temperatures as well as another mutation at another point on the genome that inhibits the production of luecine. By using polyethylene glycol (PEG), lithium acetate (LiAc), and a single stranded carrier DNA (from salmon sperm) it is possible to introduce two different plasmids (pPGK2E and pPGK2ECDC42) into the mutated yeast strain. The results of this transformation is complementation that allows both strains to grow on plates lacking luecine (Leu–) and one strain to grow at a non-permissive temperature (37°) while the other die.
Materials and Methods:
There was a deviation from the protocol that was outlined in the lab (Burke, et al 2000). Instead of having three plates, we only had two due to the lack of available plates. We obtained our data for the water control plate from the class. This procedure also called for the use of the sterile bead technique to plate the control and each transformation. Each plate contained SC-Leu– for the purpose of selecting only the cells that took in the plasmids to grow. This media was used to demonstrate the ability of the transformed yeast to grow on Lue – plates at a permissive temperature, since one of the mutations that this strain of yeast has is the inability to make its own leucine. The media, through the water control plate, shows that the original strain is in fact Lue –. To test for temperature sensitivity, the plates must be replicated using a replicating block from VWR Scientific Company and grown at a non-permissive and permissive temperatures.
Results:
|
Water control |
pPGK2ECDC42 |
pPGK2E |
|
0 |
720 |
1348 |
Table 1. Transformation of Yeast Results. The plates were made by using the sterile bead technique and allowed to grow for three days at 30°. This data shows that the strain of yeast did in fact take in the pPGK2ECDC42 and pPGK2E plasmids. It also shows that the original strain cannot grow on plates lacking leucine.
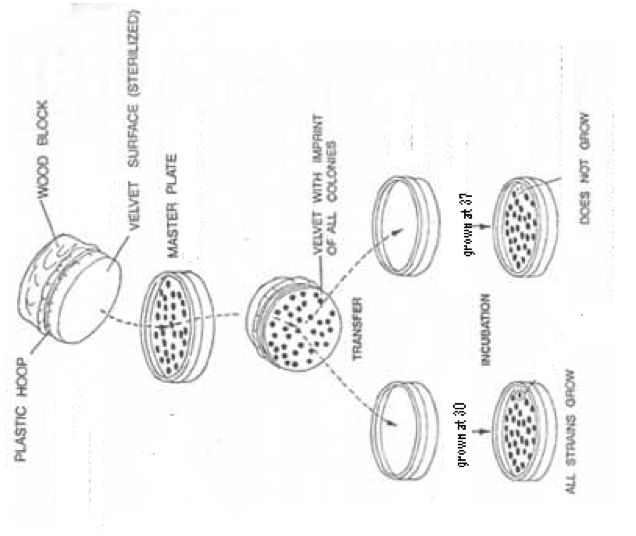
Figure 1. Technique for Replicating Plates. This diagram demonstrates how to perform the technique of replica plating. This is done for each of the two strains of transformed yeast. The first plate to be replicated must be the one to be exposed to 37°.This diagram pictures the pPGK2E strain. This strain is temperature sensitive and cannot grow at the non-permissive temperature of 37°.
|
Temp. |
pPGK2ECDC42 |
pPGK2E |
|
30° |
+ |
+ |
|
37° |
+ |
– |
Table 2. Replica Plate Results. The replicated plates were grown for three days at two temperatures: at a non-permissive 37° and at a permissive 30°. The results above show that the pPGK2E lacks the ability to grow at higher temperatures, while pPGk2ECDC42 has the ability.
The data in Table 1 and Table 2 show the complementation that has occurred between the mutant cdc42ts yeast strain and the two plasmids. Table 1 shows that both of the plasmids contain the ability to produce leucine, allowing them to grow on the Leu– plates. Table 2 shows that only the pPGK2ECDC42 complements the mutant yeast, giving it the ability to grow at the non-permissive temperature while the pPGK2E lacks this ability.
Discussion:
The first part of this lab was to show the uptake of the plasmid DNA by a cdc42ts strain of yeast. This strain is mutated so that it will die at non-permissive temperatures. Lithium acetate, PEG, and single-stranded DNA were all used and played an important role in transporting the plasmid DNA into the yeast cells. LiAc is used to puncture the yeasts’ cell wall to allow the plasmid to move into the cell. PEG has two roles. First it helps protect the cells from being over punctured by the LiAc. Secondly, and more importantly, it forces the plasmid DNA to enter the cell through the openings. The single–stranded carrier DNA also helps to carry the plasmid across the opening. The results of this first part of the experiment showed complementation occurring. The plasmid DNA complemented the yeast cell, making it capable of producing its own leucine thus growth was seen on the SB-Lue– plate. Showing that both strains grow on the Leu– plates, it is possible to now determine any temperature sensitivity that remains. Since the original strain had a mutation on CDC42 that caused it to be temperature sensitive, any growth at the non-permissive temperature would be proof of complementation. The results confirmed this. At the non-permissive temperature, the pPGK2E strain died while the pPGK2ECDC42 experienced growth.
The results of this lab show, through transformation, that complementation has occurred between the introduced plasmids and the yeasts’ own DNA.
References:
Burke, Dan, et al. Methods in Yeast Genetics. Cold Spring Harbor Laboratory Press. 2000 edition.
Chapoel, J., Johnson, J.F., Ito, Y., Ross, F.P., Zhao, H., Zheng, Y., Zou, W. (2010). Cdc42 regulates bone modeling and remodeling in mice by modulating RANKL/M-CSF signaling and osteoclast polarization. JCI 120.6, (no page number given).
Ivics, Z., Izsvák, Z., Hackett, P., and Plasterk, R. (1997). Molecular Reconstruction of Sleeping
Beauty, a Tc1-lik transposon from fish, and its transposition in human cells. Cell 91, 501-510