Microbiota of the Skin
By: Jacob Lucht
Purpose: The purpose of this experiment was to determine the type or types of bacteria living in each person’s skin by taking a sample from the nose and plating it on a selective, differential media to culture it and then using polymerase chain reactions to amplify the DNA of the bacteria and gel electrophoresis to determine which type of bacteria was present.
Theory: The human body has a large number of bacterial species living on and within it. These bacteria are typically beneficial and work with the body. One of the most popular places for these bacteria are found on the 2m2 of skin that act as a protective covering for the human body. The skin, along with beneficial bacteria, protects potentially dangerous bacteria, fungi, and viruses from getting into the body and preventing the pathogens from growing. Many species, including Micrococcus luteus, Proppionibacterium sp., and the two Staphylococcus species: Staphylococcus aureus (SA) and Staphylococcus epidermidis (SE). SA can be differentiated from other species because of its ability to ferment mannitol and cause hemolysis when grown on blood agar. The species also produces a nuclease called nuc which can exist outside the bacteria and is thermostable. Both Staphylococci species are common causes of nosocomial infections, which are drug-resistant strains that are difficult to treat. The two most common drug-resistant strains are methicillin-resistant S. aureus also called MRSA and methicillin-resistant S. epidermidis or MRSE. These species are methicillin resistant because they produce β-lactamase encoded by the blaZ gene and are able to resist the β-lactams that the antibiotic methicillin utilizes to fight the infections. They also have two other mechanisms of resistance; one is the penicillin-binding protein PBP2a which is encoded by mecA and the other is the synthesis of aminoglycoside modifying enzymes (AMEs) which are the factors involved in resistance with aminoglycosides in staphylococci.
Two types of media were used in this experiment: MSA and blood agar. MSA stands for Mannitol salt agar, which is both a differential and selective media. The selective portion is the high salt content which most other bacterial species cannot survive in. The differential, the mannitol, allows us to distinguish between the two species of Staphylococcus: SE appear as small, circular, white colonies, while SA appear yellow in color and are also circular. Staphylococcus aureus also has the ability to ferment mannitol which means will change the medium from red to yellow because the pH indicator phenol red is in the medium. The second type of media used in this experiment is blood agar media, which acts as a differential medium. Staphylococcus aureus will hemolyze the blood while Staphylococcus epidermidis will show no change when plated on blood agar.
In order to accurately identify the bacterial species present in the skin, it was necessary to extract the DNA and amplify it using polymerase chain reaction. To extract the DNA from the bacteria, restriction enzymes are used to cut the DNA at specific areas called restriction sites. Restriction enzymes are produced by bacterial species as a means of defense against bacteriophages. The enzymes are 4 to 8 base pairs long cleave the DNA on both strands to create restriction fragments. These fragments can then be seen using gel electrophoresis, which allows us to distinguish between fragments based on their size. Before injecting the DNA into the agarose gel, it must first be amplified using the polymerase chain reaction.
This reaction allows us to take a small amount of template strand and amplify a specific piece of that DNA that allows us to identify the species by the specific sequence. This process begins by denaturing the DNA into two separate strands by heating it to 94°C. The second step is the annealing of the oligonucleotide primers, which bind to complementary sequences of the single-stranded DNA at each end. For this step to occur the temperature must be brought down to somewhere between 50 and 65°C so the primers can bind to the strands of DNA. The third and final step is elongation in which a DNA polymerase (usually TAQ polymerase which is stable at high temperatures) synthesizes a complementary strand of DNA template to make a new double-stranded piece of DNA. The PCR cycle typically goes between 25 to 40 cycles with each cycle creating twice as many copies of the fragment of DNA.
Method
The experiment began by swabbing the inside of our nose and plate it onto a mannitol salt agar plate. The plate was then incubated and given to us during the next lab. Five to ten colonies were then removed from the plate with a toothpick and placed on blood agar plates and allowed to incubate. Then we removed another colony from the MSA plate and set up two PCR reactions. This began by suspending the colony in 50ul of water within an Eppendorf Tube and incubating the tube by placing in boiling water within a 400 ml beaker with foil covering the top. This was done for approximately five minutes and then immediately after we centrifuged the tube at 1300 RPM for approximately ten minutes. Next we obtained two PCR tubes, one was set up with SE primer set and the other was either SA specific primer set if the reaction was positive for mannitol or with the nuc gene if it was negative for mannitol. The following amounts were put into each tube; 7 μl of the supernatant fluid from the centrifuged tubed, 12.5 μl of 2X Mastermix that we had sit on ice and 5.5 μl of the appropriate primer mix. The next step was to amplify the mixture. This was achieved by denaturing for eight minutes at 95°C then one minute at 94°C next 30 seconds at 51 – 54°C and finally 90 seconds at 72°C. Lastly we prepared the 2% agarose gel. This followed by loading the 100 base pair ladder in well #1 and loading the PCR products in the following wells. Finally we ran the gel electrophoresis at 120 V for 40-45 minutes and checked the results under a UV illuminator.
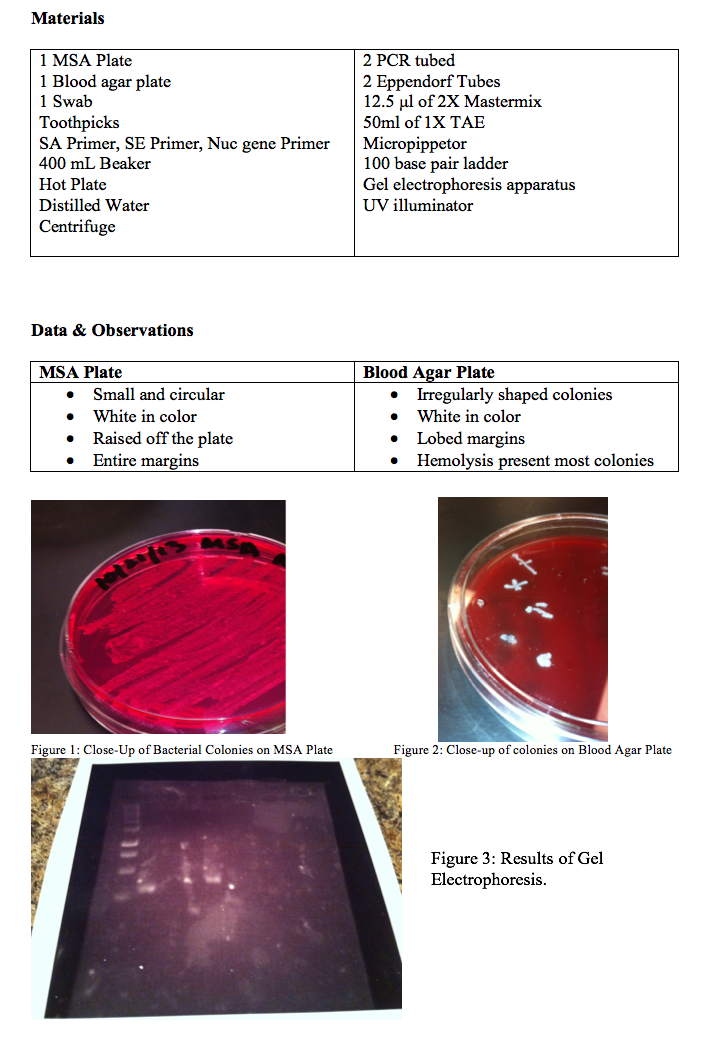
Figure 3 is the result of the gel electrophoresis of 3 different samples from nose swabs of three different students. Well 1 has the one hundred base pair ladder. It goes from 1000 base pairs at the top, to 100 base pairs at the bottom. Wells 4 and 6 had my bacterial sample mixed with SA and SE primer respectively. Wells 5-8 were bacterial samples from two other students with SE primer in well 4, nuc primer in well 5, SA primer in well 6, and SE primer in well 7.
Discussion
The results of the MSA plate indicated that the bacterial species swabbed from the individual’s nose was Staphylococcus epidermidis. This conclusion was reached because the colonies were white in appearance and there was no color change in the medium indicating that the mannitol was not fermented. The lack of fermentation led me to believe that SE was present on the plate rather than SA.
The results of the blood agar plate, however, indicated the opposite. Most of the colonies showed alpha-hemolysis of the blood which indicates that Staphylococcus aureus was the bacteria present in the skin. From this result it was thought that there was a mixture of both SE and SA because most of the colonies effectively hemolyzed the blood on the plate.
At this point, all evidence indicated that the nasal swab had a mixture of both SE and SA. Next, the DNA samples were loaded in a gel with a 100 base pair ladder and electrophoresis was performed. The loading dye unfortunately did not work. This may have happened because the loading dye wasn’t properly made or because when the loading dye was inserted into the gel, we slightly punctured the gel with the pipettor. Thus we didn’t have anything to compare our results to, so we were not able to identify for sure what bacteria was in the nose of the student.
Overall I was able to learn the steps and skills necessary to identify microorganisms from the human skin. I also learned the importance of creating a ladder correctly.
References
Scheurle, Daniela. General Microbiology Laboratory Manual. 3e. Florida Atlantic University: 2010. Pg 157-158.
Skin Microbiota Handout. Pages 1-3.
